Getting started¶
A quick overview of FLEXPART data¶
reflexible was originally developed for working with FLEXPART V8.x which has some fairly new features to how the output data is created. The latest version of FLEXPART also has functionality for saving directly to Netcdf. The ability to read this data directly is forthcoming, but for now reflexible still only works with the raw unformatted binary Fortran data FLEXPART has traditionally used for output. See the documents for information regarding FLEXPART .
A users guide for FLEXPART is available which explains the model output.
Note If you are interested in contributing functionality for other FLEXPART versions, please contact me.
reflexible was originally released as ‘pflexpart’, but as the goal is to be more generic, the package was renamed. The current release is still focused on FLEXPART, but some generalizations are starting to make their way into the code base.
reflexible is undergoing constant modifications and is not particularly stable or backward compatible code. I am trying to move in the right direction, and have moved the code now to github.org. If you are interested in contributing, feel free to contact me: John F. Burkhart
Example data¶
Several example simulations are available for testing. The simulations contain a simple backward run case and a forward case, and are suitable for testing some of the unique functions of reflexible for analysis and creation of the retroplumes.
The data is distributed with the repository:
$ ls reflexible/reflexible/uio_examples
Converting FLEXPART output to netCDF4 format¶
Reflexible is using a netCDF4 internally for doing its analysis and plotting duties. Not all the example data sets contain netcdf output, and several FLEXPART users are not yet using this functionality. This section demonstrates how to convert the FLEXPART output to netCDF4 format. In order to do that the create_ncfile script will be invoked. This script is copied into a directory in your path when reflexible is installed, so you should not worry about copying it manually.
The example data we will use is in a directory named uio_examplesBwd1_V9.02. It contains the result of processing a simple backward run case with FLEXPART. Next we can execute the fprun script:
$ fprun Bwd1_V9.02/pathnames
UserWarning: NetCDF4 files not found in output directory 'Bwd1_V9.02/outputs'.
You can always generate them from data there with the `create_ncfile` command line utility.
Read b'FLEXPART V9.0' Header: Bwd1_V9.02/outputs/header
Flexpart('Bwd1_V9.02/pathnames', nested=False)
note that you pass the pathnames of a FLEXPART run. The pathnames file has a simple structure. For example, in our case it goes like this:
/site/opt/flexpart/9.02/examples/Bwd1/options/
./outputs/
/site/opt/flexpart/WIND_FIELDS/AVAILABLE_ECMWF_EI_fields_global
============================================
So, basically in the first line indicates the <options> directory for the FLEXPART run, whereas the second line specifies the <output> directory. With this, you can easily mix and match different <options> and <output> directories for your analysis.
If we want to select the nested data instead, we can look instead at the Fwd1_V9.02/pathnames simulation with the -n flag:
$ fprun -n Fwd2_V9.02/pathnames
UserWarning: NetCDF4 files not found in output directory 'Fwd2_V9.02/outputs'.
You can always generate them from data there with the `create_ncfile` command line utility.
Read b'FLEXPART V8.2' Header: Fwd2_V9.02/outputs/header_nest
Flexpart('Fwd2_V9.02/', nested=True)
And if you want to get some info on the COMMANDS file:
$ fprun -n -C Fwd2_V9.02/pathnames
Read b'FLEXPART V8.2' Header: Fwd2_V9.02/outputs/header_nest
Flexpart('Fwd2_V9.02/pathnames', nested=True)
Command: OrderedDict([('AVG_CNC_INT', 3600), ('AVG_CNC_TAVG', 3600), ('CNC_SAMP_TIME', 300), ('CTL', 3.0), ('IFINE', 4), ('IFLUX', 0), ('IND_RECEPTOR', 1), ('IND_SOURCE', 1), ('IOUT', 5), ('IPIN', 0), ('IPOUT', 0), ('LAGESPECTRA', 0), ('LCONVECTION', 0), ('LINIT_COND', 2), ('LSUBGRID', 1), ('MDOMAINFILL', 0), ('MQUASILAG', 0), ('NESTED_OUTPUT', 1), ('OUTPUTFOREACHRELEASE', 0), ('SIM_DIR', 1), ('SIM_END', ['20070122', '180000']), ('SIM_START', ['20070121', '090000']), ('SYNC', 300), ('T_PARTSPLIT', 999999999)])
You can get more info on the supported flags by passing the -h flag to the fprun command line utility:
$ fprun -h
usage: fprun [-h] [-n] [-C] [-R] [-S] [-H HEADER_KEY] [-K] [pathnames]
positional arguments:
pathnames The Flexpart pathnames file stating where options and
output are. If you pass a dir, a 'pathnames' file will
be appended automatically. If not found yet, a FP
output dir is assumed.
optional arguments:
-h, --help show this help message and exit
-n, --nested Use a nested output.
-C, --command Print the COMMAND contents.
-R, --releases Print the RELEASES contents.
-S, --species Print the SPECIES contents.
-H HEADER_KEY, --header-key HEADER_KEY
Print the contents of H[HEADER_KEY].
-K, --header-keys Print all the HEADER keys.
Reading data out of a FLEXPART run¶
Newer versions of FLEXPART can generate convenient NetCDF4 files as output, so let’s have a quick glimpse on how you can access the different data on it.
Note In case you have a FLEXPART output that is not in NetCDF4 format, you can always make use the create_ncfile command line utility.
Open the file and print meta-information for the run:
In [1]: from netCDF4 import Dataset
In [2]: rootgrp = Dataset('./Fwd1_V9.02/outputs/grid_conc_2007012190000_nest.nc', 'r')
In [3]: print(rootgrp)
<class 'netCDF4._netCDF4.Dataset'>
root group (NETCDF4 data model, file format HDF5):
Conventions: CF-1.6
title: FLEXPART model output
institution: NILU
source: V8 model output
history: 2016-10-28 16:43 NA created by faltet on faltet-Latitude-E6430
references: Stohl et al., Atmos. Chem. Phys., 2005, doi:10.5194/acp-5-2461-200
outlon0: 1.0
outlat0: 39.5
dxout: 0.019999999553
dyout: 0.019999999553
ldirect: 1
ibdate: 20070121
ibtime: 100000
iedate: 20070122
ietime: 180000
loutstep: 3600
loutaver: 3600
loutsample: 300
lsubgrid: 1
lconvection: 0
ind_source: 1
ind_receptor: 1
itsplit: 999999999
linit_cond: 2
lsynctime: 300
ctl: 3.0
ifine: 4
iout: 5
ipout: 0
lagespectra: 0
ipin: 0
ioutputforeachrelease: 0
iflux: 0
mdomainfill: 0
mquasilag: 0
nested_output: 1
surf_only: 0
dimensions(sizes): time(33), longitude(220), latitude(220), height(1), numspec(1), pointspec(1), nageclass(1), nchar(45), numpoint(1)
variables(dimensions): int32 time(time), float32 longitude(longitude), float32 latitude(latitude), float32 height(height), <class 'str'> RELCOM(numpoint), float32 RELLNG1(numpoint), float32 RELLNG2(numpoint), float32 RELLAT1(numpoint), float32 RELLAT2(numpoint), float32 RELZZ1(numpoint), float32 RELZZ2(numpoint), int32 RELKINDZ(numpoint), int32 RELSTART(numpoint), int32 RELEND(numpoint), int32 RELPART(numpoint), float32 RELXMASS(numspec,numpoint), int32 LAGE(nageclass), int32 ORO(latitude,longitude), float32 spec001_mr(nageclass,pointspec,time,height,latitude,longitude), float32 WD_spec001(nageclass,pointspec,time,latitude,longitude), float32 DD_spec001(nageclass,pointspec,time,latitude,longitude)
groups:
We can get the info for a specific attribute just by referencing it like this:
In [4]: print(rootgrp.loutstep)
3600
We can have a look at the different dimensions and variables in the file:
In [5]: print(rootgrp.dimensions)
OrderedDict([('time', <class 'netCDF4._netCDF4.Dimension'> (unlimited): name = 'time', size = 33
), ('longitude', <class 'netCDF4._netCDF4.Dimension'>: name = 'longitude', size = 220
), ('latitude', <class 'netCDF4._netCDF4.Dimension'>: name = 'latitude', size = 220
), ('height', <class 'netCDF4._netCDF4.Dimension'>: name = 'height', size = 1
), ('numspec', <class 'netCDF4._netCDF4.Dimension'>: name = 'numspec', size = 1
), ('pointspec', <class 'netCDF4._netCDF4.Dimension'>: name = 'pointspec', size = 1
), ('nageclass', <class 'netCDF4._netCDF4.Dimension'>: name = 'nageclass', size = 1
), ('nchar', <class 'netCDF4._netCDF4.Dimension'>: name = 'nchar', size = 45
), ('numpoint', <class 'netCDF4._netCDF4.Dimension'>: name = 'numpoint', size = 1
)])
In [6]: rootgrp.variables.keys()
Out[11]: odict_keys(['time', 'longitude', 'latitude', 'height', 'RELCOM', 'RELLNG1', 'RELLNG2', 'RELLAT1', 'RELLAT2', 'RELZZ1', 'RELZZ2', 'RELKINDZ', 'RELSTART', 'RELEND', 'RELPART', 'RELXMASS', 'LAGE', 'ORO', 'spec001_mr'])
The netCDF4 Python wrappers allows to easily slice and dice variables:
In [15]: longitude = rootgrp.variables['longitude']
In [16]: print(longitude)
<class 'netCDF4._netCDF4.Variable'>
float32 longitude(longitude)
long_name: longitude in degree east
axis: Lon
units: degrees_east
standard_name: grid_longitude
description: grid cell centers
unlimited dimensions:
current shape = (220,)
filling on, default _FillValue of 9.969209968386869e+36 used
We see that ‘longitude’ is a unidimensional variable with shape (220,). Let’s read just the 10 first elements:
In [20]: longitude[:10]
Out[20]:
array([ 1.00999999, 1.02999997, 1.04999995, 1.07000005, 1.09000003,
1.11000001, 1.13 , 1.14999998, 1.16999996, 1.18999994], dtype=float32)
As only the 10 first elements are brought into memory, that permits to reduce your memory needs for your analysis.
Also, what you get from slicing netCDF4 variables are always NumPy arrays:
In [21]: type(longitude[:10])
Out[21]: numpy.ndarray
which, besides of being memory-efficient, they are what you normally use in your analysis tasks.
Also, each variable can have attached different attributes meant to add more information about what they are about:
In [23]: longitude.ncattrs()
Out[23]: ['long_name', 'axis', 'units', 'standard_name', 'description']
In [24]: longitude.long_name
Out[24]: u'longitude in degree east'
In [25]: longitude.units
Out[25]: u'degrees_east'
That’s is basically all you need to know to access the on-disk data. Feel free to play a bit more with the netCDF4 interface, because you will find it very convenient when combined with reflexible.
Basic reflexible functionality¶
Once you have checked out the code and have a sufficient FLEXPART dataset to work with you can begin to use the module. The first step is to load the package. Depending on how you checked out the code, you can accomplish this in a few different way, but the preferred is as follows:
In [1]: import reflexible as rf
The next step is to create the accessor to the FLEXPART run. In this first example we’ll work with a Forward simulation. This is easier, conceptually and a common FLEXPART use case. You pass the location of the ‘pathnames’ file to the Flexpart constructor:
In [2]: fprun = rf.Flexpart("Fwd2_V9.02/pathnames")
In [3]: type(fprun)
Out[3]: reflexible.flexpart.Flexpart
So, fprun is an instance of the Flexpart class that allows you to easily access different parts of the FLEXPART run. For example, we can access the COMMAND like this:
In [4]: fprun.Command
Out[4]:
OrderedDict([('AVG_CNC_INT', 3600),
('AVG_CNC_TAVG', 3600),
('CNC_SAMP_TIME', 300),
('CTL', 3.0),
('IFINE', 4),
('IFLUX', 0),
('IND_RECEPTOR', 1),
('IND_SOURCE', 1),
('IOUT', 5),
('IPIN', 0),
('IPOUT', 0),
('LAGESPECTRA', 0),
('LCONVECTION', 0),
('LINIT_COND', 2),
('LSUBGRID', 1),
('MDOMAINFILL', 0),
('MQUASILAG', 0),
('NESTED_OUTPUT', 1),
('OUTPUTFOREACHRELEASE', 0),
('SIM_DIR', 1),
('SIM_END', ['20070122', '180000']),
('SIM_START', ['20070121', '090000']),
('SYNC', 300),
('T_PARTSPLIT', 999999999)])
the SPECIES:
In [5]: fprun.Species
Out[5]:
defaultdict(list,
{'decay': [-999.9],
'dquer': [-9.9],
'dryvel': [-9.99],
'dsigma': [-9.9],
'f0': [-9.9],
'henry': [-9.9],
'kao': [-99.99],
'ohreact': [-9.9e-09],
'reldiff': [-9.9],
'spec_ass': [-9],
'weightmolar': [350.0],
'weta': [-9.9e-09],
'wetb': [-9.9e-09]})
But perhaps the most important accessor is the Header:
In [8]: H = fprun.Header
In [9]: type(H)
Out[9]: reflexible.data_structures.Header
Now we have a variable ‘H’ which has all the information about the run that is available from the header file. This ‘Header’ is a class instance, so the first step may be to explore some of the attributes:
In [12]: print(H.keys())
['C', 'FD', 'Heightnn', 'ORO', 'absolute_path', 'alt_unit', 'area', 'available_dates', 'available_dates_dt', 'direction', 'dxout', 'dyout', 'fill_grids', 'fp_path', 'ibdate', 'ibtime', 'iedate', 'ietime', 'ind_receptor', 'ind_source', 'iout', 'ireleaseend', 'ireleasestart', 'latitude', 'lconvection', 'ldirect', 'longitude', 'loutaver', 'loutsample', 'loutstep', 'lsubgrid', 'nageclass', 'nc', 'ncfile', 'nested', 'nspec', 'numageclasses', 'numpoint', 'numpointspec', 'numxgrid', 'numygrid', 'numzgrid', 'options', 'outheight', 'outlat0', 'outlon0', 'output_unit', 'pointspec', 'releaseend', 'releasestart', 'releasetimes', 'species', 'zpoint1', 'zpoint2']
Working with rflexible in depth¶
Assuming the above steps worked out, then we can proceed to understand the tools in a bit more detail.
Okay, let’s take a look at the example code above line by line. The first line imports the module, giving it a namespace “rf” – this is the preferred approach.
The next line creates a “fprun” instance of Flexpart,
by passing the pathnames of a FLEXPART run.:
In [24]: fprun = rf.Flexpart("Fwd2_V9.02/pathnames")
and from there, we can easily have access to the Header container:
In [25]: H = fprun.Header
The Header is central to reflexible. This contains much information about the FLEXPART run, and enable plotting, labeling of plots, looking up dates of runs, coordinates for mapping, etc. All this information is contained in the Header. See for example:
In [27]: print(dir(H))
['C', 'FD', 'Heightnn', 'ORO', '__class__', '__delattr__', '__dict__', '__dir__', '__doc__', '__eq__', '__format__', '__ge__', '__getattribute__', '__getitem__', '__gt__', '__hash__', '__init__', '__le__', '__lt__', '__module__', '__ne__', '__new__', '__reduce__', '__reduce_ex__', '__repr__', '__setattr__', '__sizeof__', '__str__', '__subclasshook__', '__weakref__', '_gridarea', 'absolute_path', 'add_trajectory', 'alt_unit', 'area', 'available_dates', 'available_dates_dt', 'direction', 'dxout', 'dyout', 'fill_grids', 'fp_path', 'ibdate', 'ibtime', 'iedate', 'ietime', 'ind_receptor', 'ind_source', 'iout', 'ireleaseend', 'ireleasestart', 'keys', 'latitude', 'lconvection', 'ldirect', 'longitude', 'loutaver', 'loutsample', 'loutstep', 'lsubgrid', 'nageclass', 'nc', 'ncfile', 'nested', 'nspec', 'numageclasses', 'numpoint', 'numpointspec', 'numxgrid', 'numygrid', 'numzgrid', 'options', 'outheight', 'outlat0', 'outlon0', 'output_unit', 'pointspec', 'releaseend', 'releasestart', 'releasetimes', 'species', 'zpoint1', 'zpoint2']
This will show you all the attributes associated with the Header.
H is now an object in your workspace. Using Ipython you can explore the methods and attributes of H.
Data Containers: H.FD and H.C¶
There are a couple attributes of the Header that contain data. These are important to understand in order to work with reflexible.
Reasonably, you should now want to read in some of the data from your run. At this point, the Header or, H variable, should now have an attribute ‘FD’ which is again a dictionary of the FLEXPART grids (think ‘Flexpart Data’):
In [13]: H.FD
Out[13]: <reflexible.data_structures.FD at 0x7f83bc4c5898>
In [15]: H.FD.keys()[:3] # for only the 3 first entries
Out[15]: [(0, '20070121100000'), (0, '20070121110000'), (0, '20070121120000')]
Look at the keys of the dictionary to see what information is stored. The actual data is keyed by tuples: (nspec, datestr) where nspec is the species number and datestr is a YYYYMMDDHHMMSS string for the grid timestep:
In [16]: fd = H.FD[(0, '20070121100000')]
In [17]: fd.data_cube.shape
Out[18]: (60, 40, 1, 1, 1)
And we can see what the dimensions are as they relate to the shape:
In [19]: fd.data_cube.dims
Out[20]: ('longitude', 'latitude', 'height', 'pointspec', 'nageclass')
Some information about the individual data for the timestep is provided:
In [33]: fd.keys()
Out[33]:
['data_cube',
'gridfile',
'itime',
'timestamp',
'species',
'rel_i',
'spec_i',
'dry',
'wet',
'slabs',
'shape',
'max',
'min']
These are available as attributes of the flexpart data class:
In [97]: fd.timestamp
Out[97]: datetime.datetime(2007, 1, 21, 10, 0)
In [102]: fd.species
Out[102]: ['TRACER']
There is a second attribute, H.C, that is meant to provide ‘Cumulative` sensitivity at each time step, as well as some cumulative values such as a total_column attribute.
H.C is similar to the H.FD object described above, but contains the Cumulative sensitivity at each time step, so you can use it for plotting retroplumes.
It is important to understand the differences between H.FD and H.C while working with reflexible. If we look closely at the keys of H.FD:
In [29]: H.FD.keys()[:3]
Out[29]: [(0, '20070121100000'), (0, '20070121110000'), (0, '20070121120000')]
You’ll see that the dictionary is primary keyed by a set of tuples.
These tuples represent (s, date), where s is the specied ID and
date is the date of a grid in FLEXPART.
However, if we look at the keys of the H.C dictionary:
In [30]: H.C.keys()[:3]
Out[30]: [(0, 0)]
We see only tuples, now keyed by (s, rel_id), where s is still the species ID, but rel_id is the release ID. These release IDs correspond to the times in H.releasetimes which is a list of the release times. In a forward run, this will be the same. However, in a backward run, there will be differences.
Each tuple is a key to another dictionary, that contains the data. Currently there are differences between the way the data is stored in H.FD and in H.C, but future versions are working to make these two data stores common.
So we know now H.C is keyed by (s,k) where s is an integer for the species #, and k is an integer for the release id.
Working with Backward Simulations¶
The biggest difference for the H.C object is when you have a backward simulation. First, for a backward simulation, the attribute will not exist unless you explicitly call for it:
In [135]: bwrun = rf.Flexpart('Bwd1_V9.02/pathnames')
/Data/johnbur/.conda/envs/shyft/lib/python3.6/site-packages/reflexible-0.5.0-py3.6.egg/reflexible/flexpart.py:45: UserWarning: NetCDF4 files not found in output directory 'Bwd1_V9.02/outputs'. You can always generate them from data there with the `create_ncfile` command line utility.
self.fp_output))
Read b'FLEXPART V9.0' Header: Bwd1_V9.02/outputs/header
In [136]: bwH = bwrun.Header
In [137]: bwH.C
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
<ipython-input-137-71db37c7653c> in <module>()
----> 1 bwH.C
~/.conda/envs/shyft/lib/python3.6/site-packages/reflexible-0.5.0-py3.6.egg/reflexible/conv2netcdf4/legacy_structures.py in __getattr__(self, attr)
178 if attr == "__getstate__":
179 return lambda: None
--> 180 return self[attr]
181
182 def __setattr__(self, attr, value):
KeyError: 'C'
Because reading all the data for a backward run is time and memory intensive, it will not be done automatically. Instead, you need to explicitly ask for it:
In [138]: bwH.fill_backward()
getting grid for: ['20070121090000', '20070121100000', '20070121110000', '20070121120000', '20070121130000', '20070121140000', '20070121150000', '20070121160000', '20070121170000', '20070121180000', '20070121190000', '20070121200000', '20070121210000', '20070121220000', '20070121230000', '20070122000000', '20070122010000', '20070122020000', '20070122030000', '20070122040000', '20070122050000', '20070122060000', '20070122070000', '20070122080000', '20070122090000', '20070122100000', '20070122110000', '20070122120000', '20070122130000', '20070122140000', '20070122150000', '20070122160000', '20070122170000']
Assumed V8 Flexpart
Using readgrid from FortFlex
60 40 1 [0] 0 0 33 1
20070121090000
20070121100000
20070121110000
20070121120000
20070121130000
20070121140000
20070121150000
20070121160000
20070121170000
20070121180000
20070121190000
20070121200000
20070121210000
20070121220000
20070121230000
20070122000000
20070122010000
20070122020000
20070122030000
20070122040000
20070122050000
20070122060000
20070122070000
20070122080000
20070122090000
20070122100000
20070122110000
20070122120000
20070122130000
20070122140000
20070122150000
20070122160000
20070122170000
[0]
At which point, reflexible will run through all the releasetimes and calculate the cumulative sensitivity so that retroplumes may be calculated.
Note the shapes of the data returned when running backward simulations different:
In [154]: bw_fd = bwH.FD[(0, '20070121090000')]
In [155]: c = bwH.C[(0,0)]
In [156]: bw_fd.shape
Out[156]: (60, 40, 1, 1, 1)
In [157]: c.shape
Out[157]: (60, 40, 1)
Adding Trajectories¶
I use the read_trajectories() function to read the trajectories.txt
file and get the trajectories from the run output directory.:
T = rf.read_trajectories(H)
Note, that the only required parameter is the Header “H”, this provides all the metadata for the function to read the trajectories. This is a function that accepts simply the “H” instance or a path to a trajectories file.
Now we can see how we might batch process a backward run and create
total column plots as well as add the trajectory information to the
plots. The following lines plot the data sets using the
plot_totalcolumn(), plot_trajectory(), and
plot_footprint().
Warning
There is a lot of reliance on the mapping module in the plot_routines. If you
have problems, see the mapping.py file. Or the mapping
docstrings. Documentation of this module is presently incomplete but I
am working on it.
In order to reuse figures which is much faster when working with the basemap module, I create a “None” objects for passing the figure instances around:
TC = None
After that we loop over the keys (s=species, and k=rel_i) of the H.C
attribute we created by calling fill_backward. Note, I named this
attribute C for “Cumulative”. In each iteration, for a new
combination of s,k we pull the data object out of the dictionary. The
“data” object is returned from the function readgridV8() and has
some attributes that we can use later in conjunction with the
plot_totalcolumn() function and for saving and naming the
figures. See for example the following lines:
for s, k in H.C:
data = H.C[(s, k)]
TC = rf.plot_totalcolumn(H, data, map_region='Europe', FIGURE=TC)
TC = rf.plot_trajectory(H, T, k, FIGURE=TC)
filename = '%s_tc_%s.png' % (data.species, data.timestamp)
TC.fig.savefig(filename)
This will create filenames based on the data metadata and save the figure to the path defined by filename. You should now have several images looking like this:
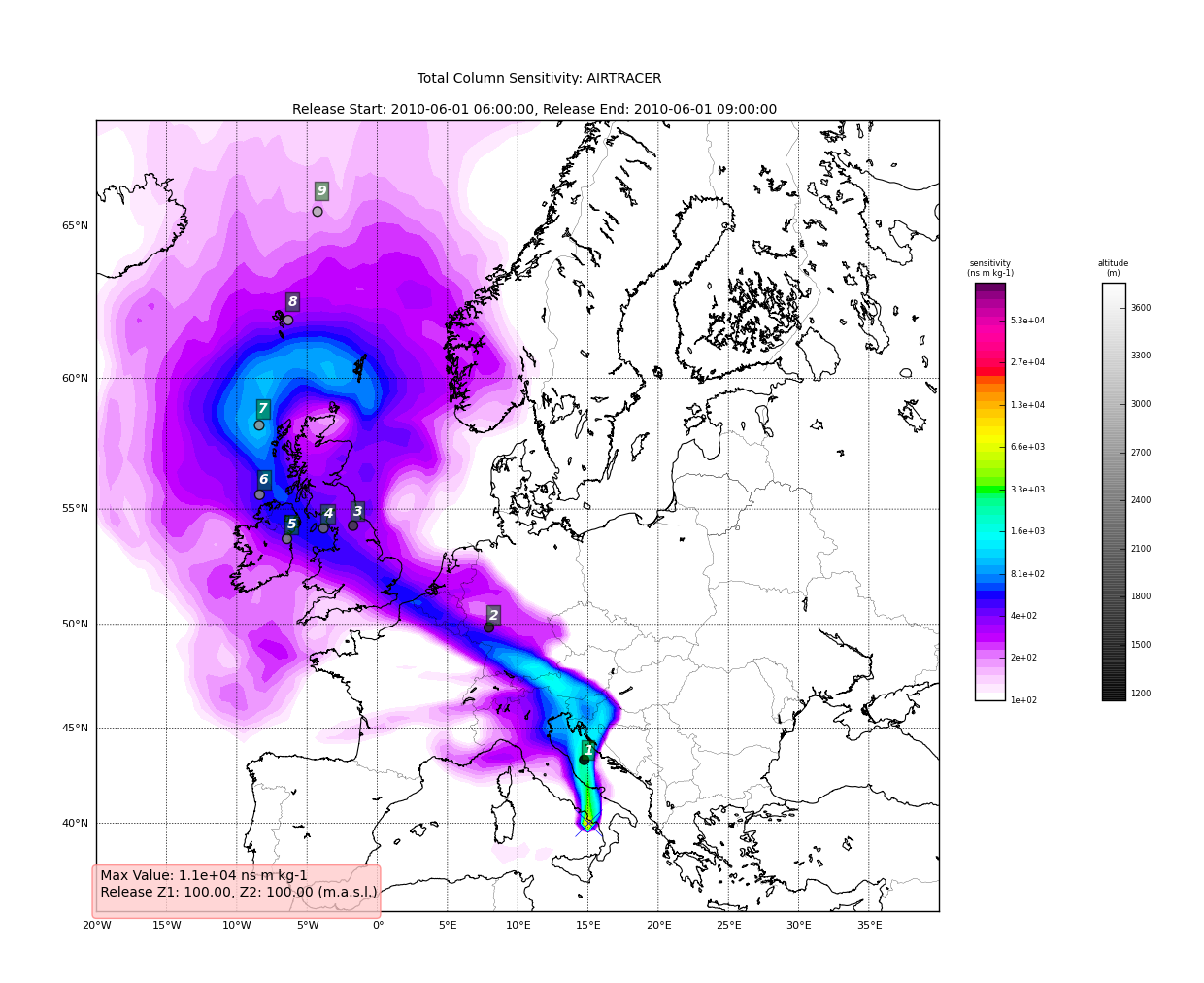
The next step is the use the source and learn more about the functionality of the module. I highly recommend the Ipython interpreter and use of the Tab key to explore the modules methods.
Enjoy!